Vivlion’s Technology
PRCISRTM CRISPR gRNA libraries empower next-generation CRISPR screens
A PCR- and cloning-free production process
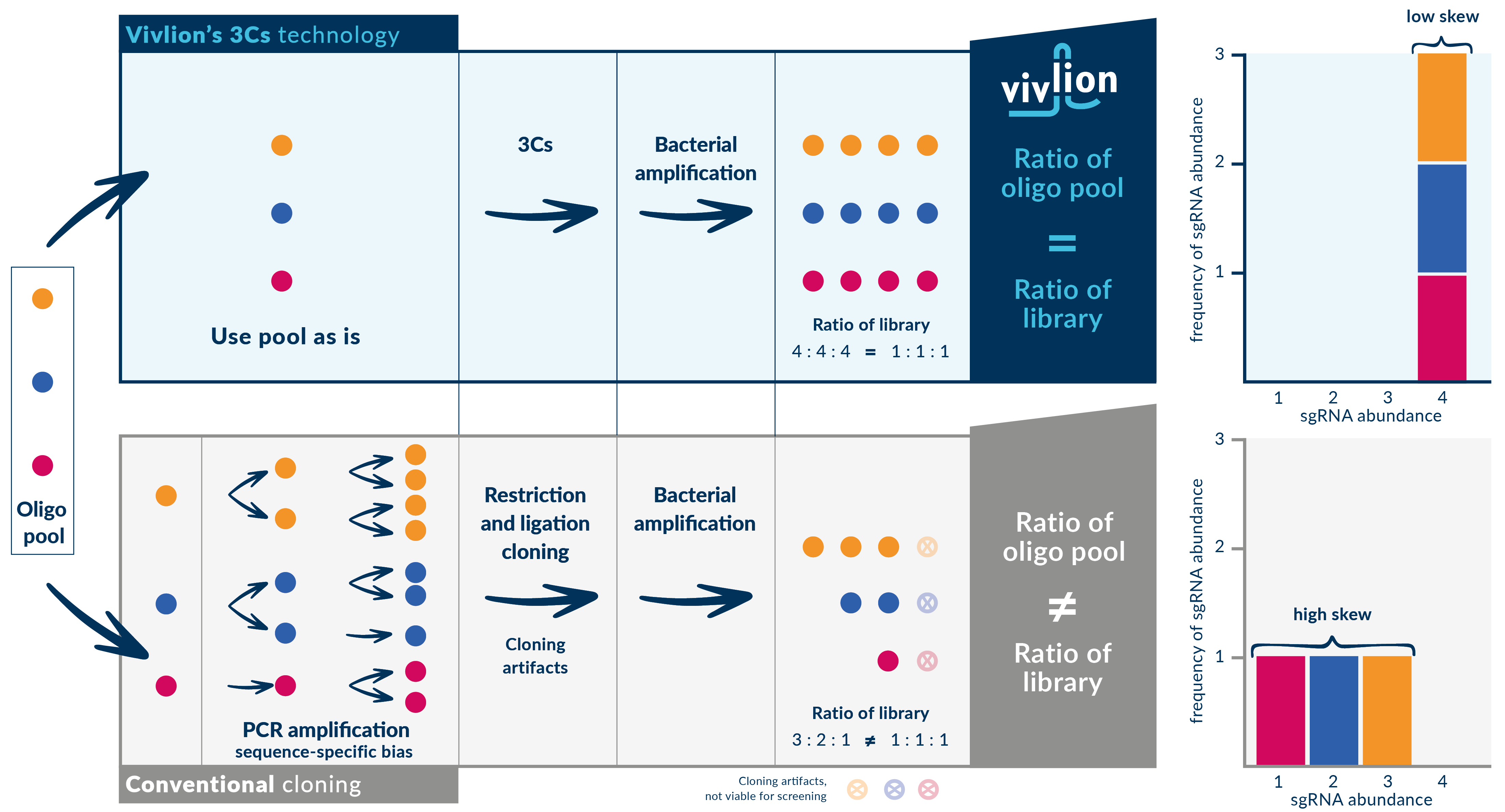
PRCISR™ CRISPR libraries are manufactured using our proprietary Covalently-Closed-Circular-synthesized (3Cs) technology which overcomes major challenges encountered in conventional CRISPR library production. Conventional library production relies on PCR amplification of oligonucleotide pools, a step that introduces bias and results in incomplete libraries and in a non-uniform sgRNA representation. Compensating for a non-uniform CRISPR library may require up to 1,000-fold coverage to detect low-abundance guides. This means that CRISPR screens can become prohibitively large.
In contrast, the 3Cs CRISPR library production process employed by Vivlion, preserves the uniform distribution of the original oligo pool in the final library. This allows for a much smaller experiment without the risk of missing potential hits while maintaining or even improving data quality, as most pertinently demonstrated with Vivlion’s Pergamon single targeting library.
-Our workflow comprises the following steps:
Oligonucleotide annealing: sgRNA-encoding oligonucleotides are annealed to circular, deoxyuridine (dU)-containing single-stranded PRCISR™ CRISPR template plasmids.
Polymerase-mediated extension: The annealed oligonucleotides are extended by a polymerase, forming hetero-duplexed double-stranded PRCISR™ CRISPR plasmids. Library diversity is solely defined by the number of sgRNA-encoding oligonucleotides, ensuring a precise and diverse sgRNA library.
Bacterial amplification: The plasmids are amplified in bacteria, removing the template plasmids and yielding the final PRCISR™ CRISPR sgRNA library.
With our 3Cs technology, library complexity is not a limiting factor anymore. Therefore, Vivlion can offer combinatorial libraries in multiplex and pre-defined fixed-pair sgRNA designs.
Pre-defined fixed-pair sgRNA libraries such as Vivlion’s fixed-pair Alexandria library, in which both sgRNAs are targeting the same gene, improve editing efficiency compared to single-targeting approaches.
Multiplex libraries in which both sgRNAs are targeting different loci allow testing all combinations of two sgRNA pools and enable novel applications like DNA region excision.
How superior uniformity and low skew are achieved
Vivlion’s proprietary workflow utilizes the oligonucleotide pool directly for the 3Cs reaction, followed by bacterial amplification, without relying on a PCR-based amplification step, as shown in the image below. The ratio of sgRNAs in the oligonucleotide pool equals the one in the library. Different sgRNAs of the library show the same abundance, leading to a low distribution skew of the library.
A histogram plot of sgRNA sequences in the final library shows the sgRNA distribution:
The x-axis represents sgRNA abundance and the y-axis indicates the number of sgRNAs at a specific abundance level. Ideally, all sgRNA sequences are equally abundant.
Challenges of conventional CRISPR library generation
PCR amplification bias: Conventional cloning requires the PCR amplification of an oligonucleotide pool. During each cycle, certain sgRNA sequences are amplified more than others, leading to under-representation and over-representation (“sequence-specific bias”). The polymerase error rate introduces mutations in the amplified PCR product, affecting the final sgRNA sequences.
Cloning inefficiencies: Post-PCR, the oligo pool is incorporated into plasmids using restriction and ligation cloning. This process is plagued by inefficiencies, such as vector ligation and re-ligation issues (“cloning artifacts”).
Bacterial amplification skew: The cloned library is amplified in bacteria to obtain sufficient plasmid DNA. However, sequence-specific bias and cloning artifacts lead to a high skew, compromising library uniformity.
The PRCISR™ CRISPR advantage
Direct oligo pool utilization: Vivlion’s 3Cs technology utilizes the oligonucleotide pool directly, avoiding sequence-specific PCR bias.
Cloning artifact elimination: 3Cs technology bypasses restriction-/ligation-based cloning, eliminating cloning artifacts.
Highly efficient amplification: Libraries are amplified in bacteria using highly efficient protocols, producing the final PRCISR™ CRISPR library with low skew, accurately reflecting the distribution of the original oligonucleotide pool
By choosing Vivlion’s 3Cs technology, you ensure that your CRISPR sgRNA libraries are free from the biases and artifacts that compromise conventional methods. Experience unparalleled uniformity and accuracy in your genetic research.
For more details on how Vivlion’s 3Cs technology can enhance your CRISPR experiments, contact us today and request our comprehensive data sheets and handling protocols.
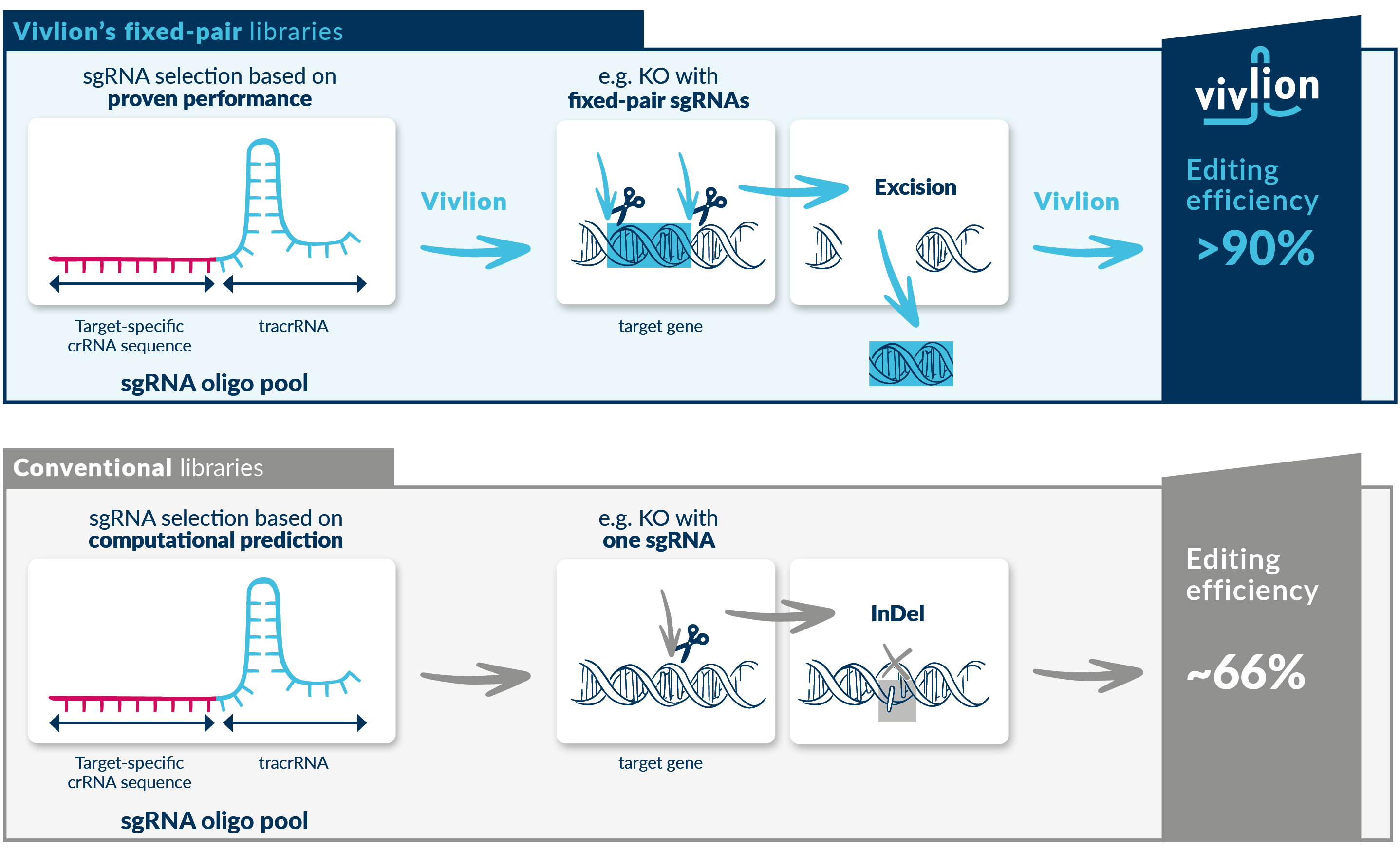
How editing efficiency is increased
The sgRNAs selected for many conventional CRISPR libraries are typically based on computational predictions of their activity. The highest scoring sgRNAs are assembled into a library that delivers a single sgRNA to a cell, resulting in a single gene editing event. For knockout-experiments, the introduction of a single insertion or deletion (InDel) leads to gene editing efficiencies of approximately 66%.
However, the sgRNAs selected for Vivlion’s PRCISR™ CRISPR fixed-pair libraries, e.g. the Alexandria library, are based on published data of hundreds of CRISPR screens. From this data, sgRNAs are selected and assembled into a library of pre-defined pairs, delivering two highly functional sgRNAs to each cell, each targeting a gene of interest. The sgRNA pairs are designed to excise and remove a part of the target gene, thereby improving gene editing efficiencies to more than 90%.
Very excited to continue our productive relationship with Vivlion GmbH. It has been a great experience working with their talented team and excellent gene editing reagents!
Contact us
We would like to hear more about your CRISPR questions and applications – please reach out to us by completing the following form: