PRCISR™ CRISPR
Vivlion’s CRISPR-enabled Discovery Platform
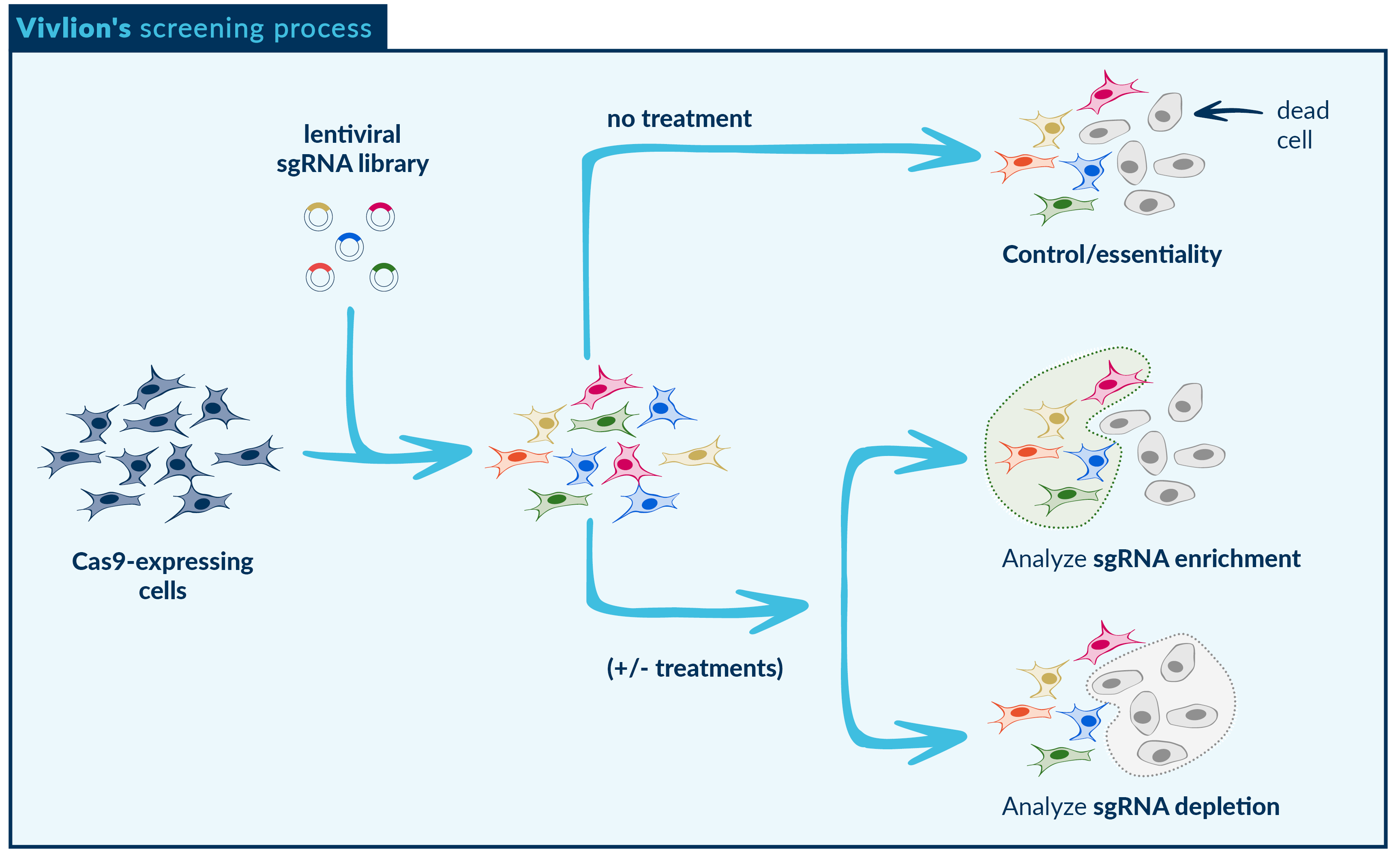
End-to-end PRCISR™ CRISPR screening
We provide tailored CRISPR screening and bioinformatic analysis services to meet your projects specific needs. Utilizing our proprietary PRCISR™ CRISPR technology with single or combinatorial libraries, we ensure precise and reliable results. Our expert team offers detailed consultation for your experimental strategy and design, ensuring optimal outcomes. We engineer the cells of your choice, conduct the screen, perform the bioinformatic analysis, and deliver the results to you.
Communication is of the utmost importance to us; therefore, a dedicated scientific project manager will be in contact with you throughout the duration of the project.
Discover potential read-outs for your CRISPR screen
Functional screening with Vivlion
Analysis based on cellular viability
Viability assays separate cells based on whether they proliferate or survive when knocked out. These screens identify genes that influence cell fitness or, in case of combinatorial libraries, gene dependencies. When viability screens are conducted in the presence of a certain selective pressure, such as a drug, they can uncover the genetic response to that specific stimuli.
Positive selection: identifies cells with knockouts that provide a survival or growth advantage. These cells are enriched during a screen and sequenced to identify the gene perturbations associated with survival. These types of screens are often used for identifying mutations that confer drug resistance.
Negative selection: identifies genes that confer a growth or survival disadvantage. In these screens, referred to as dropout screens, the genes of interest will be depleted and inferred comparing the surviving cells to the initial population. Negative selection is often used to identify gene essentiality and dependencies, such as synthetic lethal genes, as well as mutations that enhance drug activity.
Flow cytometry-based analysis
Fluorescence-activated cell sorting (FACS) can be used to physically sort the edited cells into subpopulations, and is therefore suitable for phenotypic screening that does not involve cell proliferation.
For cell surface/biomarker analysis edited cells can be first stained with a fluorescent antibody specific for a biomarker of interest and then FACS is used to sort the cells based on the presence or absence of the marker.
Reporter genes, such as those for fluorescence or luminescence, can be inserted to signal pathway activity or the expression of specific genes. Cells can then be sorted by FACS based on reporter presence or activity changes.
These assays can be used to investigate the effect of gene perturbations on cell phenotypes, pathways, or proteins of interest.
Analysis of gene expression (single-cell RNA sequencing)
In complex screens, single-cell RNA sequencing (scRNA-seq) is used to profile transcriptomic changes across individual cells in response to different sgRNA modifications.
This allows for a high-resolution read-out of gene expression changes and the identification of gene networks and pathways influenced by specific knockouts.
Planning your screening project
Count on us to meticulously plan your CRISPR screening project. We ensure that every critical parameter is tailored to your needs, leveraging our expertise to deliver precise and reliable results.
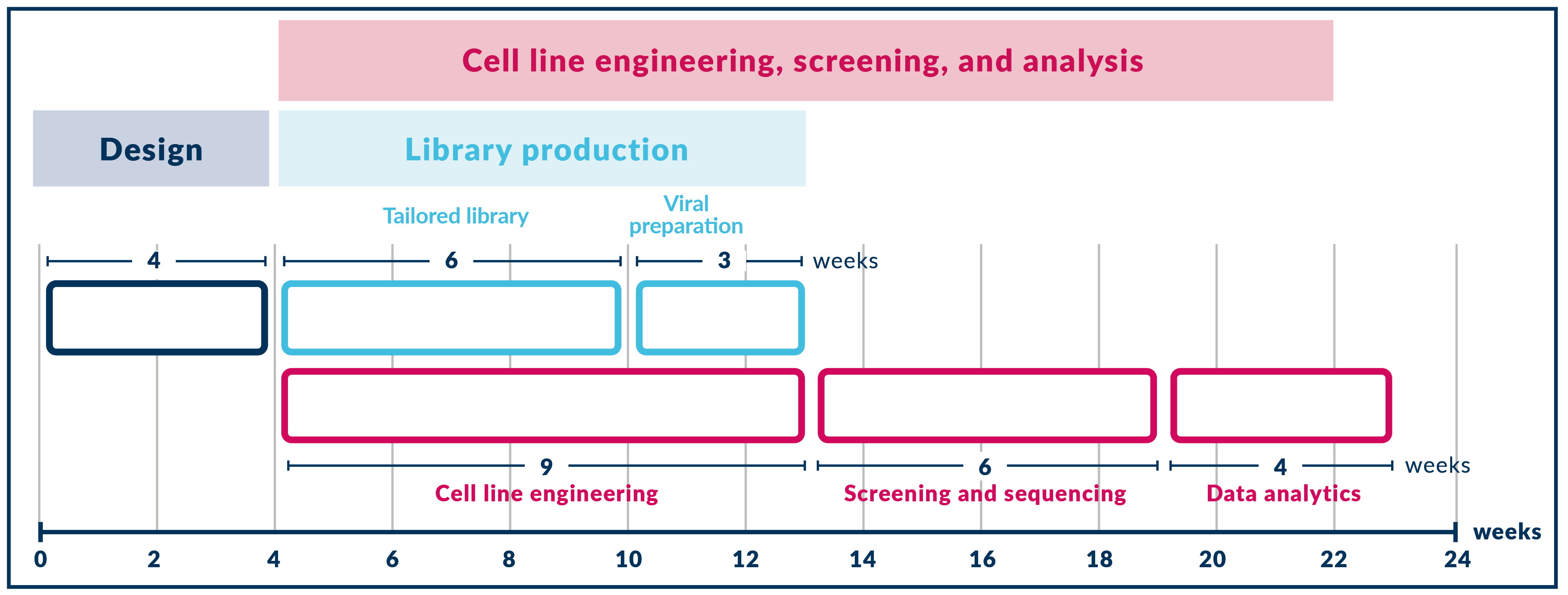
Our PRCISR™ screening services are organized into four distinct phases. By following these phases, we ensure your CRISPR screening project is efficiently executed and delivered on time, providing you with high-quality, reliable results. Partner with us for a seamless, expert-driven CRISPR screening experience!
Phase 1 – Experimental design
- Planning of critical parameters of the experimental setup such as cellular context, treatments applied, control concept, read-out, and duration
- Library design and format: choose between our PRCISRTM CRISPR off-the-shelf libraries or a PRCISRTM CRISPR library specifically tailored to your experiment
Phase 2 – Preparatory work
- Production of a tailored PRCISRTM CRISPR library (not applicable if an off-the-shelf PRCISRTM CRISPR library is chosen)
- Engineering and testing Cas9 in cell line(s) of choice
- Cell line quality control (QC) to confirm identity and test for contamination
- Transfer of materials such as client compounds (if needed)
Phase 3 – Screening experiment
- Expansion of cells to the required scale
- Performance of screen
- Next-generation sequencing (NGS) of samples
Phase 4 – Bioinformatic analysis
- Processing of NGS and screening samples
- Hit-calling utilizing the agreed pipelines
- Transfer of data and final report
Our basic analysis package includes NGS quality control and phenotypic hit calling. For deeper insights, we offer optional data integration to enrich the resulting experimental data.
Comprehensive analysis packages
Ready to start your CRISPR screening project?
Reach out via our contact form to start planning your project. We will then together discuss the parameters for library design and desired screen outcomes for your experiments, ensuring we meet your specific needs and goals.
Contact us
We would like to hear more about your CRISPR questions and applications – please reach out to us by completing the following form: